Modern Kernel Methods
in Machine Learning:
Part II
Danica J. Sutherland(she/her)
Computer Science, University of British Columbia
ETICS "summer" school, Oct 2022
(Swipe or arrow keys to move through slides; for a menu to jump; to show more.)
Yesterday, we saw:
- RKHS is a function space,
- Reproducing property:
- Representer theorem:
- Can use to do kernel ridge regression, SVMs, etc
Today:
- Kernel mean embeddings of distributions
- Gaussian processes and probabilistic numerics
- Kernel approximations, for better computation
- Neural tangent kernels
Mean embeddings of distributions
- Represent point as :
- Represent distribution as :
- Last step assumed e.g.
- Okay. Why?
- One reason: ML on distributions [Szabó+ JMLR-16]
- More common reason: comparing distributions
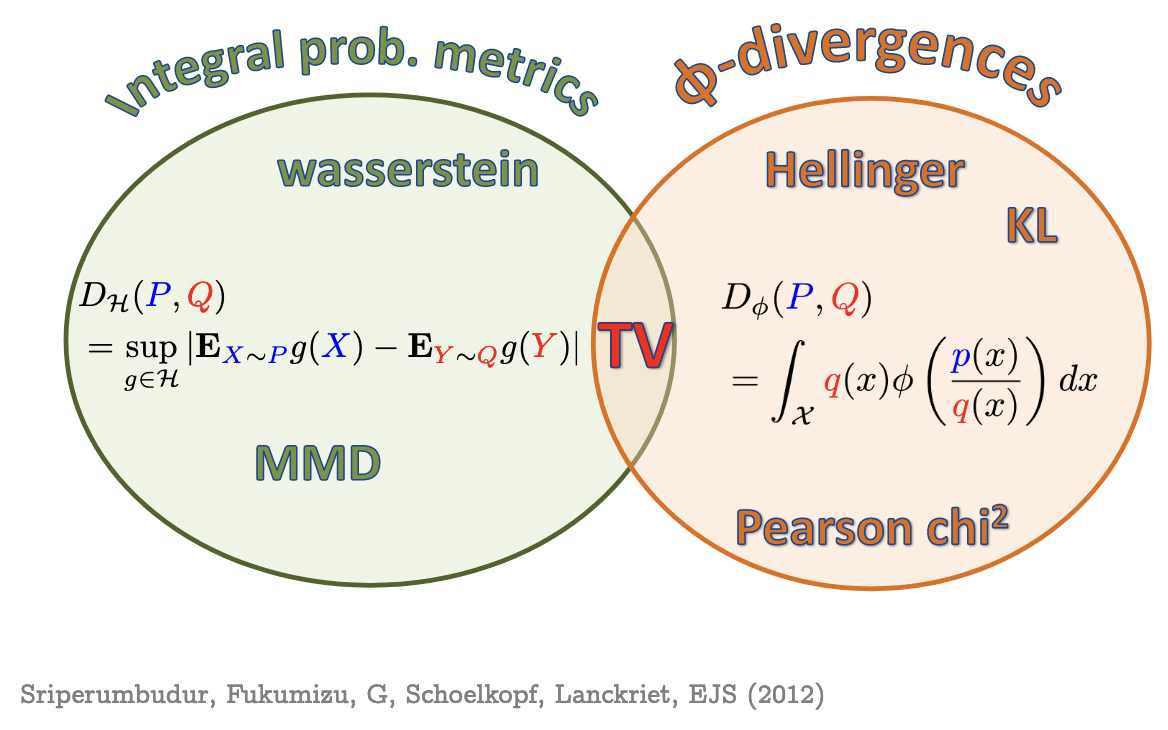
Maximum Mean Discrepancy
- Last line is Integral Probability Metric (IPM) form
- is called “witness function” or “critic”: high on , low on
MMD properties
- , symmetry, triangle inequality
- If is characteristic, then iff
- i.e. is injective
- Makes MMD a metric on probability distributions
- Universal => characteristic
- Linear kernel: is just Euclidean distance between means
Application: Kernel Herding
- Want a "super-sample" from :
- If , error
- Greedily minimize the MMD:
- Get approximation instead of with random samples
![]()
Estimating MMD from samples
MMD vs other distances
- MMD has easy estimator
- block or incomplete estimators are for , but noisier
- For bounded kernel, estimation error
- Independent of data dimension!
- But, no free lunch…the value of the MMD generally shrinks with growing dimension, so constant error gets worse relatively
![]()
Application: Two-sample testing
- Given samples from two unknown distributions
- Question: is ?
- Hypothesis testing approach:
- Reject if
- Do smokers/non-smokers get different cancers?
- Do Brits have the same friend network types as Americans?
- When does my laser agree with the one on Mars?
- Are storms in the 2000s different from storms in the 1800s?
- Does presence of this protein affect DNA binding? [MMDiff2]
- Do these dob and birthday columns mean the same thing?
- Does my generative model match ?
What's a hypothesis test again?
MMD-based testing
- : converges in distribution to…something
- Infinite mixture of s, params depend on and
- Can estimate threshold with permutation testing
- : asymptotically normal
- Any characteristic kernel gives consistent test…eventually
- Need enormous if kernel is bad for problem
Classifier two-sample tests
![]()
- is the accuracy of on the test set
- Under , classification impossible:
- With where ,
get
Deep learning and deep kernels
- is one form of deep kernel
- Deep models are usually of the form
- With a learned
- If we fix , have with
- Same idea as NNGP approximation
- Generalize to a deep kernel:
Normal deep learning deep kernels
- Take
- Final function in will be
- With logistic loss: this is Platt scaling
![]()
“Normal deep learning deep kernels” – so?
- This definitely does not say that deep learning is (even approximately) a kernel method
- …despite what some people might want you to think
![]()
- We know theoretically deep learning can learn some things faster than any kernel method [see Malach+ ICML-21 + refs]
- But deep kernel learning ≠ traditional kernel models
- exactly like how usual deep learning ≠ linear models
Optimizing power of MMD tests
- Asymptotics of give us immediately that
, , are constants:
first term usually dominates
- Pick to maximize an estimate of
- Use from before, get from U-statistic theory
- Can show uniform convergence of estimator
- Get better tests (even after data splitting)
Application: (S)MMD GANs
- An implicit generative model:
- A generator net outputs samples from
- Minimize estimate of on a minibatch
- MMD GAN:
- SMMD GAN:
- Scaled MMD uses kernel properties to ensure smooth loss for by making witness function smooth [Arbel+ NeurIPS-18]
- Uses
- Standard WGAN-GP better thought of in kernel framework
Application: distribution regression/classification/…
![]()
Independence
- iff for all measurable ,
- Let's implement for RKHS functions , :where is
Cross-covariance operator and independence
- If , then
- If ,
- If , are characteristic:
- implies [Szabó/Sriperumbudur JMLR-18]
- iff
- iff (sum squared singular values)
- HSIC: "Hilbert-Schmidt Independence Criterion"
HSIC
- Linear case: is cross-covariance matrix,
HSIC is squared Frobenius norm - Default estimator (biased, but simple):
where
![]()
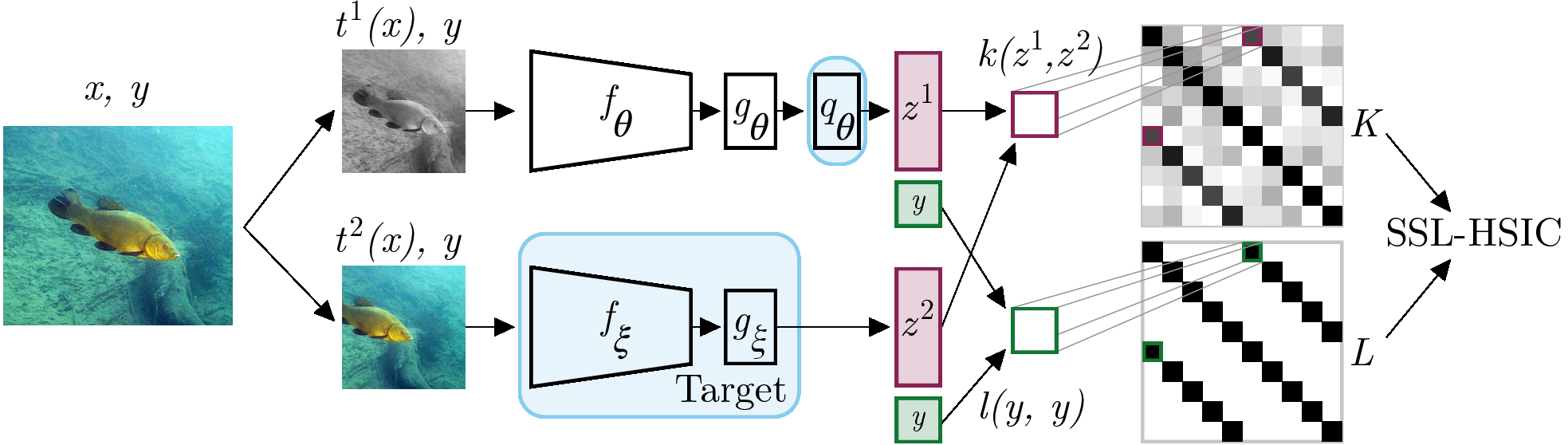
- Maximizes dependence between image features and its identity on a minibatch
- Using a learned deep kernel based on
Recap
- Mean embedding
- is 0 iff (for characteristic kernels)
- is 0 iff (for characteristic , )
- After break: last interactive session exploring testing
- More details:


















![]()
 ,
,  ,
,  ,
,  ,
,